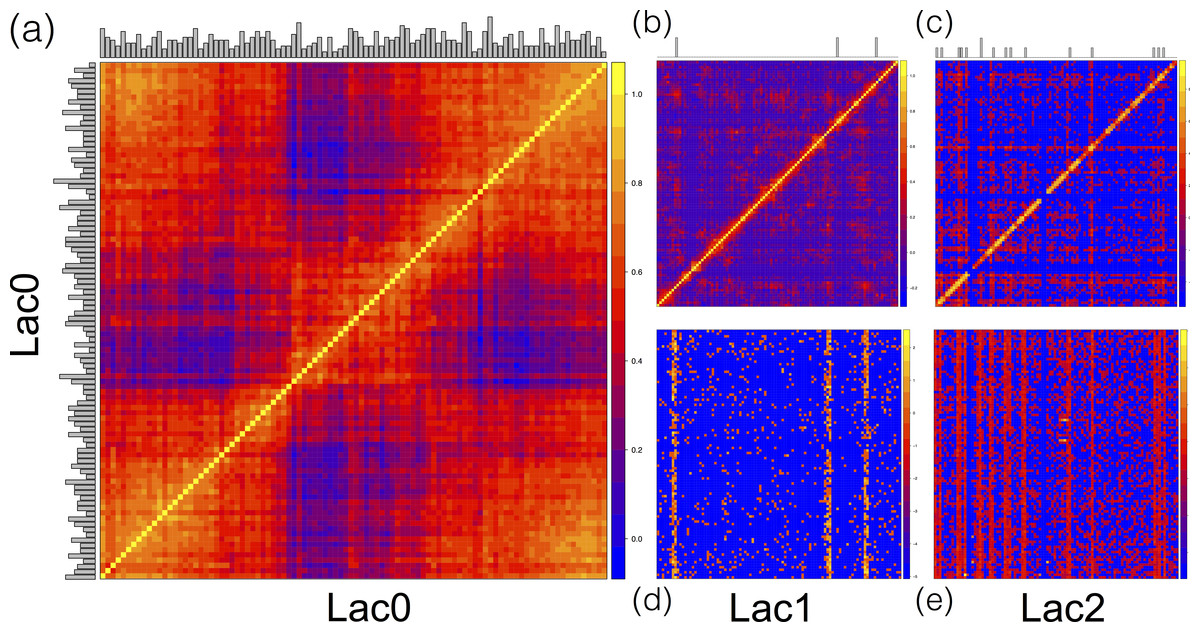
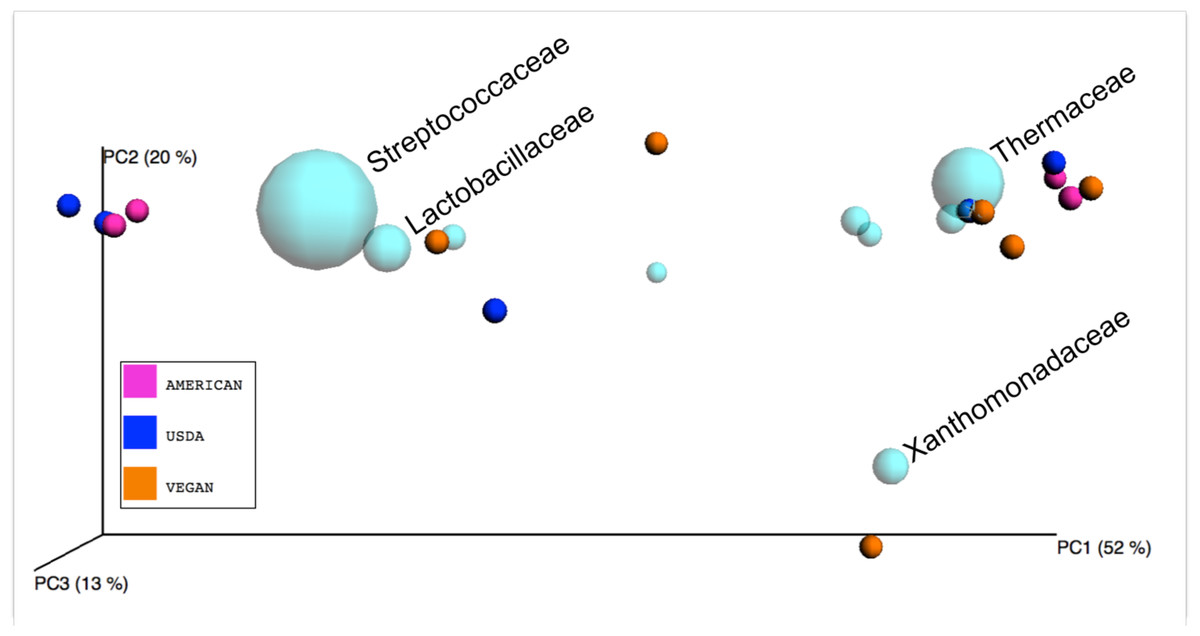
Well, just got pointed to this paper:
Metagenomic chromosome conformation capture (meta3C) unveils the diversity of chromosome organization in microorganisms | eLife by Martial Marbouty, Axel Cournac, Jean-François Flot, Hervé Marie-Nelly, Julien Mozziconacci, Romain Koszul. Seems potentially really interesting.
It is similar in concept and in many aspects to a paper we published in PeerJ earlier in the year (see
Beitel et al., 2014 Beitel CW, Froenicke L, Lang JM, Korf IF, Michelmore RW, Eisen JA, Darling AE. (2014) Strain- and plasmid-level deconvolution of a synthetic metagenome by sequencing proximity ligation products. PeerJ 2:e415 http://dx.doi.org/10.7717/peerj.415.
Yet despite the similarities to our paper and to another paper that was formally published around the time of ours, this new paper does not mention these other pieces of work any where in the introduction as having any type of "prior work" relevance. Instead, they wait until late in their discussion:
Taking advantage of chromatin conformation capture data to address genomic questions is a dynamic field: while this paper was under review, two studies were released that also aimed at exploiting the physical contacts between DNA molecules to deconvolve genomes from controlled mixes of microorganisms (Beitel et al., 2014; Burton et al., 2014).
Clearly, what they are trying to do here is to claim that since they paper was submitted before these other two (including ours) was published, that they should get some sort of "priority" for their work. Let's look at that in more detail. Their paper was received May 9, 2014. Our paper was published online May 27 and the other related paper by Burton et al. was published online May 22. In general, if a paper on what your paper is about comes out just after you submit your paper, while your paper is still in review, the common, normal thing to be asked to do is to rewrite your paper to deal with the fact that you were, in essence, scooped. But that does not really appear to be the case here. They are treating this in a way as "oh look, some new papers came out at the last minute and we have commented on them." The last minute would be in this case, 6 months before this new paper was accepted. Seems like a long time to treat this as "ooh - a new paper came up that we will add a few comments about".
But - one could quibble about the ethics and policies of dealing with papers that were published after one submitted one's own paper. From my experience, I have always had to do major rewrites to deal with such papers. But maybe E-Life has different policies. Who knows. But that is where things get really annoying here. This is because it was May 27 when our FINAL paper came out online at PeerJ. However, the
preprint of the paper was published on February 27, more than two months before their paper was even submitted. So does this mean that the authors of this new paper do not believe that preprints exist? It is pretty clear on the web site for our paper that there is a preprint that was published earlier. Given what they were working on - something directly related to what our preprint/paper was about, one would assume they would have seen it with a few simple Google searches. Or a reviewer might have pointed them to it. Maybe not. I do not know. But either way, our preprint was published long before their paper was submitted and therefore I believe they should have discussed it in more detail.
Is this a sign that some people believe preprints are nothing more than rumors? I hope not. Preprints are a great way to share research prior to the delays that can happen in peer review. And in my opinion, preprints should count as prior research and be cited as such. I note - the
Burton group in their paper in G3 also did not reference our preprint in what I consider to be a reasonable manner. They add some comments in their acknowledgements
While this manuscript was in preparation, a preprint describing a related method appeared in PeerJ PrePrints (Beitel et al. 2014a). Note added in proof: this preprint was subsequently published (Beitel et al. 2014b).
Given that our preprint was published before their paper was submitted too, I believe that they also should have made more reference to it in their paper. But again, I can only guess that both the Burton and the Marbouty group just do not see preprints as being respectable scientific objects. That is a bad precedent to set and I think the wrong one too. And it is a shame. A preprint is a publication. No - it is not peer reviewed. But that does not mean it should not be considered part of the scientific literature in some way. I note - this new paper from the Marbouty group seems really interesting. Not sure I want to dig into it any deeper if they are going to play games with the timing of submission vs. published "papers" as part of how they are positioning themselves to be viewed as doing something novel.